Overview
| Basic Information | |
|---|---|
| IMG/M Taxon OID | 3300001769 Open in IMG/M |
| GOLD Reference (Study | Sequencing Project | Analysis Project) | Gs0094242 | Gp0056359 | Ga0012999 |
| Sample Name | Hydrothermal vent plume microbial communities from the Mid Cayman Rise - Shallow Background Supr60 |
| Sequencing Status | Permanent Draft |
| Sequencing Center | |
| Published? | N |
| Use Policy | Open |
| Dataset Contents | |
|---|---|
| Total Genome Size | 109162239 |
| Sequencing Scaffolds | 21 |
| Novel Protein Genes | 22 |
| Associated Families | 22 |
| Dataset Phylogeny | |
|---|---|
| Taxonomy Groups | Number of Scaffolds |
| All Organisms → cellular organisms → Bacteria → FCB group → Candidatus Marinimicrobia | 1 |
| All Organisms → cellular organisms → Bacteria | 3 |
| All Organisms → cellular organisms → Bacteria → Proteobacteria | 1 |
| All Organisms → cellular organisms → Bacteria → FCB group → Bacteroidetes/Chlorobi group → Bacteroidetes | 1 |
| Not Available | 10 |
| All Organisms → cellular organisms → Bacteria → Proteobacteria → Alphaproteobacteria | 1 |
| All Organisms → cellular organisms → Bacteria → Proteobacteria → Alphaproteobacteria → Pelagibacterales → Pelagibacteraceae → Candidatus Pelagibacter | 1 |
| All Organisms → cellular organisms → Bacteria → Proteobacteria → Alphaproteobacteria → Rhodospirillales → unclassified Rhodospirillales → Rhodospirillales bacterium | 1 |
| All Organisms → cellular organisms → Bacteria → Proteobacteria → delta/epsilon subdivisions → Deltaproteobacteria → unclassified Deltaproteobacteria → Deltaproteobacteria bacterium | 1 |
| All Organisms → cellular organisms → Archaea | 1 |
Ecosystem and Geography
| Ecosystem Assignment (GOLD) | |
|---|---|
| Name | Hydrothermal Vent Plume Microbial Communities From The Cayman Rise, Cayman Islands |
| Type | Environmental |
| Taxonomy | Environmental → Aquatic → Marine → Hydrothermal Vents → Unclassified → Hydrothermal Vent Plume → Hydrothermal Vent Plume Microbial Communities From The Cayman Rise, Cayman Islands |
| Alternative Ecosystem Assignments | |
|---|---|
| Environment Ontology (ENVO) | marine hydrothermal vent biome → marine hydrothermal plume → hydrothermal fluid |
| Earth Microbiome Project Ontology (EMPO) | Free-living → Non-saline → Subsurface (non-saline) |
| Location Information | ||||||||
|---|---|---|---|---|---|---|---|---|
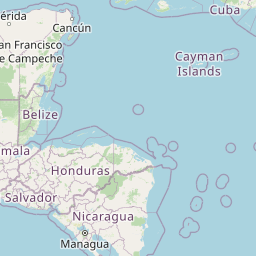
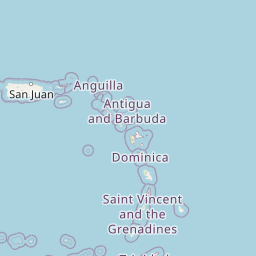
| Location | Shallow Background, Mid Cayman Rise | |||||||
| Coordinates | Lat. (o) | 18.35 | Long. (o) | -81.85 | Alt. (m) | N/A | Depth (m) | 2000 |
| Location on Map | ||||||||
|
| ||||||||
| Zoom: | Powered by OpenStreetMap © | |||||||