Overview
| Basic Information | |
|---|---|
| IMG/M Taxon OID | 3300003535 Open in IMG/M |
| GOLD Reference (Study | Sequencing Project | Analysis Project) | Gs0112330 | Gp0107643 | Ga0060329 |
| Sample Name | Petroleum reservoir microbial communities from Reconcavo Basin, Brazil, analyzing oil degradation - Bahia-well BA- 1 |
| Sequencing Status | Permanent Draft |
| Sequencing Center | |
| Published? | N |
| Use Policy | Open |
| Dataset Contents | |
|---|---|
| Total Genome Size | 88315047 |
| Sequencing Scaffolds | 11 |
| Novel Protein Genes | 13 |
| Associated Families | 13 |
| Dataset Phylogeny | |
|---|---|
| Taxonomy Groups | Number of Scaffolds |
| All Organisms → cellular organisms → Bacteria | 2 |
| Not Available | 3 |
| All Organisms → cellular organisms → Archaea → Euryarchaeota → Stenosarchaea group → Methanomicrobia → Methanotrichales → Methanotrichaceae → Methanothrix → unclassified Methanothrix → Methanothrix sp. | 1 |
| All Organisms → cellular organisms → Bacteria → Terrabacteria group → Firmicutes → Clostridia → Eubacteriales → Syntrophomonadaceae → Syntrophomonas → Syntrophomonas wolfei | 1 |
| All Organisms → cellular organisms → Archaea → Euryarchaeota → unclassified Euryarchaeota → Euryarchaeota archaeon ADurb.Bin009 | 1 |
| All Organisms → cellular organisms → Bacteria → Terrabacteria group → Chloroflexi | 1 |
| All Organisms → cellular organisms → Bacteria → Terrabacteria group → Chloroflexi → Anaerolineae → Anaerolineales → Anaerolineaceae → unclassified Anaerolineaceae → Anaerolineaceae bacterium | 1 |
| All Organisms → cellular organisms → Archaea → Euryarchaeota → Stenosarchaea group → Candidatus Methanofastidiosa → unclassified Candidatus Methanofastidiosa → Candidatus Methanofastidiosa archaeon | 1 |
Ecosystem and Geography
| Ecosystem Assignment (GOLD) | |
|---|---|
| Name | Petroleum Reservoir Microbial Communities From Reconcavo Basin, Brazil, Analyzing Oil Degradation |
| Type | Environmental |
| Taxonomy | Environmental → Terrestrial → Unclassified → Unclassified → Unclassified → Petroleum Reservoir → Petroleum Reservoir Microbial Communities From Reconcavo Basin, Brazil, Analyzing Oil Degradation |
| Alternative Ecosystem Assignments | |
|---|---|
| Environment Ontology (ENVO) | terrestrial biome → oil reservoir → oil |
| Earth Microbiome Project Ontology (EMPO) | Free-living → Non-saline → Subsurface (non-saline) |
| Location Information | ||||||||
|---|---|---|---|---|---|---|---|---|
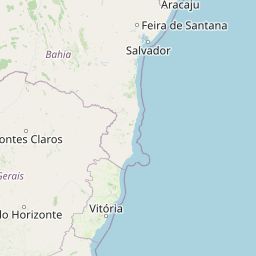
| Location | Brazil: State of Bahia | |||||||
| Coordinates | Lat. (o) | -12.2 | Long. (o) | -38.11 | Alt. (m) | N/A | Depth (m) | 500 to 1000 |
| Location on Map | ||||||||
|
| ||||||||
| Zoom: | Powered by OpenStreetMap © | |||||||