Overview
| Basic Information | |
|---|---|
| Taxon OID | 2035918001 Open in IMG/M |
| Scaffold ID | WWPM1086_GHCZ4414_g1 Open in IMG/M |
| Source Dataset Name | Activated sludge microbial communities from Commune de Morges, Switzerland - plasmid pool Visp-2009 (Newbler) |
| Source Dataset Category | Metagenome |
| Source Dataset Use Policy | Open |
| Sequencing Center | DOE Joint Genome Institute (JGI) |
| Sequencing Status | Permanent Draft |
| Scaffold Components | |
|---|---|
| Scaffold Length (bps) | 726 |
| Total Scaffold Genes | 2 (view) |
| Total Scaffold Genes with Ribosome Binding Sites (RBS) | 0 (0.00%) |
| Novel Protein Genes | 1 (view) |
| Novel Protein Genes with Ribosome Binding Sites (RBS) | 0 (0.00%) |
| Associated Families | 1 |
| Taxonomy | |
|---|---|
| All Organisms → cellular organisms → Eukaryota → Viridiplantae → Streptophyta → Streptophytina → Embryophyta → Tracheophyta → Euphyllophyta → Spermatophyta → Magnoliopsida → Mesangiospermae → eudicotyledons → Gunneridae → Pentapetalae → rosids → fabids → Malpighiales → Salicaceae → Saliceae → Populus | (Source: UniRef50) |
Ecosystem & Geography
| Source Dataset Ecosystem |
|---|
| Engineered → Wastewater → Nutrient Removal → Dissolved Organics (Aerobic) → Activated Sludge → Wastewater Plasmid Pools → Activated Sludge Microbial Communities From Commune De Morges, Switzerland |
| Source Dataset Sampling Location | ||||||||
|---|---|---|---|---|---|---|---|---|
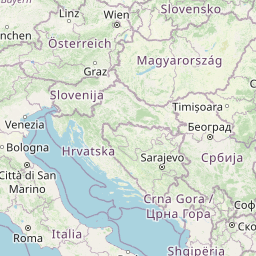
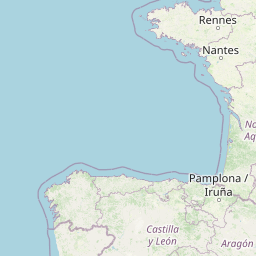
| Location Name | Visp, Switzerland | |||||||
| Coordinates | Lat. (o) | 46.30044 | Long. (o) | 7.8594 | Alt. (m) | Depth (m) | ||
| Location on Map | ||||||||
|
| ||||||||
| Zoom: | Powered by OpenStreetMap © | |||||||