Overview
| Basic Information | |
|---|---|
| IMG/M Taxon OID | 3300019080 Open in IMG/M |
| GOLD Reference (Study | Sequencing Project | Analysis Project) | Gs0117946 | Gp0217079 | Ga0193342 |
| Sample Name | Metatranscriptome of marine eukaryotic protist communities collected during Tara Oceans survey from station TARA_110 - TARA_N000001762 (ERX1782368-ERR1712081) |
| Sequencing Status | Permanent Draft |
| Sequencing Center | Canada's Michael Smith Genome Sciences Centre |
| Published? | N |
| Use Policy | Open |
| Dataset Contents | |
|---|---|
| Total Genome Size | 31767241 |
| Sequencing Scaffolds | 27 |
| Novel Protein Genes | 35 |
| Associated Families | 27 |
| Dataset Phylogeny | |
|---|---|
| Taxonomy Groups | Number of Scaffolds |
| Not Available | 17 |
| All Organisms → cellular organisms → Eukaryota → Opisthokonta → Metazoa → Eumetazoa → Bilateria → Protostomia → Ecdysozoa → Panarthropoda → Arthropoda → Mandibulata → Pancrustacea → Crustacea → Multicrustacea → Hexanauplia → Copepoda → Neocopepoda → Gymnoplea → Calanoida → Temoridae → Eurytemora → Eurytemora affinis | 8 |
| All Organisms → cellular organisms → Eukaryota → Opisthokonta | 1 |
| All Organisms → cellular organisms → Eukaryota → Opisthokonta → Metazoa → Eumetazoa → Bilateria → Protostomia → Ecdysozoa → Panarthropoda → Arthropoda → Mandibulata → Pancrustacea → Crustacea → Multicrustacea → Hexanauplia → Copepoda → Neocopepoda → Gymnoplea → Calanoida → Pseudodiaptomidae → Pseudodiaptomus → Pseudodiaptomus poplesia | 1 |
Ecosystem and Geography
| Ecosystem Assignment (GOLD) | |
|---|---|
| Name | Marine Viral And Eukaryotic Protist Communities Collected From Different Water Depths During Tara Oceans Survey |
| Type | Environmental |
| Taxonomy | Environmental → Aquatic → Marine → Unclassified → Unclassified → Marine → Marine Viral And Eukaryotic Protist Communities Collected From Different Water Depths During Tara Oceans Survey |
| Alternative Ecosystem Assignments | |
|---|---|
| Environment Ontology (ENVO) | marine biome → marine water body → sea water |
| Earth Microbiome Project Ontology (EMPO) | Free-living → Saline → Water (saline) |
| Location Information | ||||||||
|---|---|---|---|---|---|---|---|---|
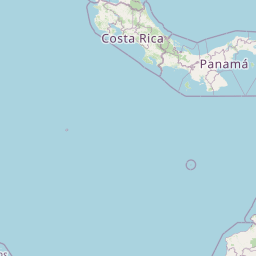
| Location | South Pacific Ocean: TARA_110 | |||||||
| Coordinates | Lat. (o) | -1.9389 | Long. (o) | -84.611 | Alt. (m) | N/A | Depth (m) | 5 |
| Location on Map | ||||||||
|
| ||||||||
| Zoom: | Powered by OpenStreetMap © | |||||||