Overview
| Basic Information | |
|---|---|
| Taxon OID | 3300031800 Open in IMG/M |
| Scaffold ID | Ga0310122_10010794 Open in IMG/M |
| Source Dataset Name | Marine microbial communities from Western Arctic Ocean, Canada - CB6_Bottom_1051 |
| Source Dataset Category | Metagenome |
| Source Dataset Use Policy | Open |
| Sequencing Center | DOE Joint Genome Institute (JGI) |
| Sequencing Status | Permanent Draft |
| Scaffold Components | |
|---|---|
| Scaffold Length (bps) | 5576 |
| Total Scaffold Genes | 9 (view) |
| Total Scaffold Genes with Ribosome Binding Sites (RBS) | 6 (66.67%) |
| Novel Protein Genes | 3 (view) |
| Novel Protein Genes with Ribosome Binding Sites (RBS) | 2 (66.67%) |
| Associated Families | 3 |
| Taxonomy | |
|---|---|
| Not Available | (Source: ) |
Ecosystem & Geography
| Source Dataset Ecosystem |
|---|
| Environmental → Aquatic → Marine → Oceanic → Unclassified → Marine → Marine Microbial Communities From Western Arctic Ocean, Canada |
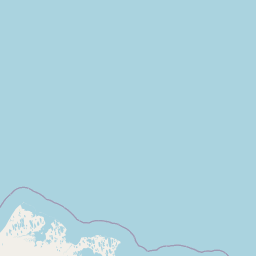
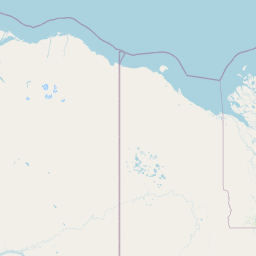
| Source Dataset Sampling Location | ||||||||
|---|---|---|---|---|---|---|---|---|
| Location Name | Canada: Western Arctic Ocean | |||||||
| Coordinates | Lat. (o) | 74.6947 | Long. (o) | -146.6612 | Alt. (m) | Depth (m) | 3678 | |
| Location on Map | ||||||||
|
| ||||||||
| Zoom: | Powered by OpenStreetMap © | |||||||