Overview
| Basic Information | |
|---|---|
| IMG/M Taxon OID | 3300029561 Open in IMG/M |
| GOLD Reference (Study | Sequencing Project | Analysis Project) | Gs0133130 | Gp0283476 | Ga0244853 |
| Sample Name | Human fecal microbial communities from Shanghai Jiao Tong University, China - SZAXPI021997-32 |
| Sequencing Status | Permanent Draft |
| Sequencing Center | Beijing Genomics Institute (BGI) |
| Published? | N |
| Use Policy | Open |
| Dataset Contents | |
|---|---|
| Total Genome Size | 168214220 |
| Sequencing Scaffolds | 6 |
| Novel Protein Genes | 7 |
| Associated Families | 7 |
| Dataset Phylogeny | |
|---|---|
| Taxonomy Groups | Number of Scaffolds |
| All Organisms → cellular organisms → Bacteria → FCB group → Bacteroidetes/Chlorobi group → Bacteroidetes → Bacteroidia → Bacteroidales → Rikenellaceae → Alistipes → Alistipes putredinis | 1 |
| All Organisms → cellular organisms → Bacteria → Terrabacteria group → Firmicutes → Clostridia → Eubacteriales → Oscillospiraceae → Faecalibacterium → Faecalibacterium prausnitzii | 1 |
| All Organisms → cellular organisms → Bacteria → Terrabacteria group → Firmicutes | 1 |
| All Organisms → cellular organisms → Bacteria → Terrabacteria group → Firmicutes → Clostridia → Eubacteriales | 1 |
| All Organisms → cellular organisms → Bacteria → Terrabacteria group → Firmicutes → Clostridia → Eubacteriales → unclassified Eubacteriales → Clostridiales bacterium | 1 |
| All Organisms → cellular organisms → Bacteria → Terrabacteria group → Firmicutes → Clostridia → unclassified Clostridia → Clostridia bacterium | 1 |
Ecosystem and Geography
| Ecosystem Assignment (GOLD) | |
|---|---|
| Name | Human Fecal Microbial Communities From Shanghai Jiao Tong University, China |
| Type | Host-Associated |
| Taxonomy | Host-Associated → Human → Digestive System → Large Intestine → Fecal → Human Fecal → Human Fecal Microbial Communities From Shanghai Jiao Tong University, China |
| Alternative Ecosystem Assignments | |
|---|---|
| Environment Ontology (ENVO) | Unclassified |
| Earth Microbiome Project Ontology (EMPO) | Host-associated → Animal → Animal distal gut |
| Location Information | ||||||||
|---|---|---|---|---|---|---|---|---|
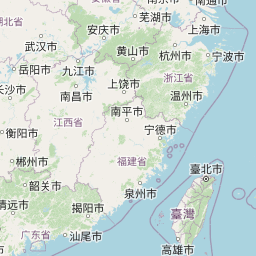
| Location | China: Shanghai | |||||||
| Coordinates | Lat. (o) | 31.2123446 | Long. (o) | 121.4684853 | Alt. (m) | N/A | Depth (m) | N/A |
| Location on Map | ||||||||
|
| ||||||||
| Zoom: | Powered by OpenStreetMap © | |||||||