Overview
| Basic Information | |
|---|---|
| IMG/M Taxon OID | 2162886015 Open in IMG/M |
| GOLD Reference (Study | Sequencing Project | Analysis Project) | Gs0046710 | Gp0052459 | Ga0010948 |
| Sample Name | Grass soil microbial communities from Rothamsted Park, UK - FO (Mercury 0.02g/kg) assembled |
| Sequencing Status | Finished |
| Sequencing Center | 454 Life Sciences |
| Published? | N |
| Use Policy | Open |
| Dataset Contents | |
|---|---|
| Total Genome Size | 50520744 |
| Sequencing Scaffolds | 29 |
| Novel Protein Genes | 29 |
| Associated Families | 29 |
| Dataset Phylogeny | |
|---|---|
| Taxonomy Groups | Number of Scaffolds |
| All Organisms → cellular organisms → Bacteria | 6 |
| All Organisms → cellular organisms → Bacteria → Acidobacteria → unclassified Acidobacteria → Acidobacteria bacterium | 4 |
| All Organisms → cellular organisms → Bacteria → Proteobacteria | 2 |
| All Organisms → cellular organisms → Bacteria → PVC group → Verrucomicrobia | 4 |
| All Organisms → cellular organisms → Bacteria → Acidobacteria → Acidobacteriia → Acidobacteriales → Acidobacteriaceae | 1 |
| All Organisms → cellular organisms → Bacteria → PVC group → Verrucomicrobia → unclassified Verrucomicrobia → Verrucomicrobia bacterium | 8 |
| Not Available | 4 |
Ecosystem and Geography
| Ecosystem Assignment (GOLD) | |
|---|---|
| Name | Grass Soil Microbial Communities From Rothamsted Park Plot 3D, Harpenden, Uk |
| Type | Environmental |
| Taxonomy | Environmental → Terrestrial → Soil → Unclassified → Unclassified → Grass Soil → Grass Soil Microbial Communities From Rothamsted Park Plot 3D, Harpenden, Uk |
| Alternative Ecosystem Assignments | |
|---|---|
| Environment Ontology (ENVO) | grassland biome → land → grassland soil |
| Earth Microbiome Project Ontology (EMPO) | Free-living → Non-saline → Soil (non-saline) |
| Location Information | ||||||||
|---|---|---|---|---|---|---|---|---|
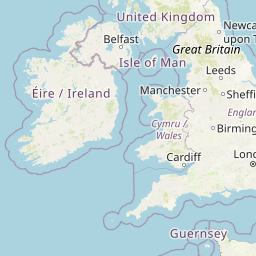
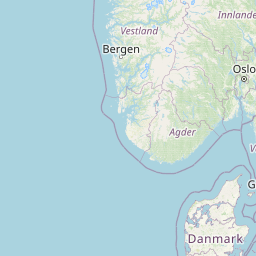
| Location | United Kingdom: Rothamsted, Harpenden | |||||||
| Coordinates | Lat. (o) | 51.804241 | Long. (o) | -0.372114 | Alt. (m) | N/A | Depth (m) | 0 to .21 |
| Location on Map | ||||||||
|
| ||||||||
| Zoom: | Powered by OpenStreetMap © | |||||||