Overview
| Basic Information | |
|---|---|
| Taxon OID | 3300034158 Open in IMG/M |
| Scaffold ID | Ga0370507_0001316 Open in IMG/M |
| Source Dataset Name | Peat soil microbial communities from wetlands in Alaska, United States - Frozen_pond_06D_18 |
| Source Dataset Category | Metagenome |
| Source Dataset Use Policy | Open |
| Sequencing Center | DOE Joint Genome Institute (JGI) |
| Sequencing Status | Permanent Draft |
| Scaffold Components | |
|---|---|
| Scaffold Length (bps) | 8843 |
| Total Scaffold Genes | 14 (view) |
| Total Scaffold Genes with Ribosome Binding Sites (RBS) | 13 (92.86%) |
| Novel Protein Genes | 2 (view) |
| Novel Protein Genes with Ribosome Binding Sites (RBS) | 2 (100.00%) |
| Associated Families | 2 |
| Taxonomy | |
|---|---|
| All Organisms → cellular organisms → Bacteria → Proteobacteria | (Source: UniRef50) |
Ecosystem & Geography
| Source Dataset Ecosystem |
|---|
| Environmental → Terrestrial → Soil → Wetlands → Unclassified → Untreated Peat Soil → Peat Soil Microbial Communities From Wetland Fen In Alaska, United States |
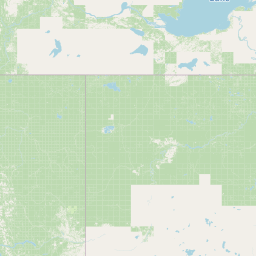
| Source Dataset Sampling Location | ||||||||
|---|---|---|---|---|---|---|---|---|
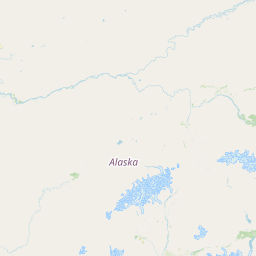
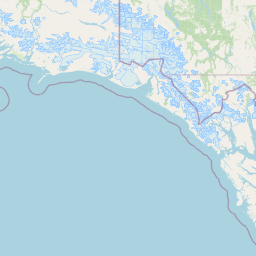
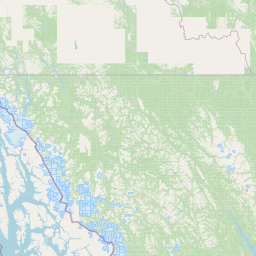
| Location Name | USA: Alaska | |||||||
| Coordinates | Lat. (o) | 64.9144 | Long. (o) | -147.8354 | Alt. (m) | Depth (m) | 0 | |
| Location on Map | ||||||||
|
| ||||||||
| Zoom: | Powered by OpenStreetMap © | |||||||