Overview
| Basic Information | |
|---|---|
| Taxon OID | 3300000558 Open in IMG/M |
| Scaffold ID | Draft_11629488 Open in IMG/M |
| Source Dataset Name | Wastewater microbial communities from Syncrude, Ft. McMurray, Alberta - West In Pit SyncrudeMLSB2011 |
| Source Dataset Category | Metagenome |
| Source Dataset Use Policy | Open |
| Sequencing Center | McGill University |
| Sequencing Status | Permanent Draft |
| Scaffold Components | |
|---|---|
| Scaffold Length (bps) | 34224 |
| Total Scaffold Genes | 51 (view) |
| Total Scaffold Genes with Ribosome Binding Sites (RBS) | 37 (72.55%) |
| Novel Protein Genes | 10 (view) |
| Novel Protein Genes with Ribosome Binding Sites (RBS) | 9 (90.00%) |
| Associated Families | 10 |
| Taxonomy | |
|---|---|
| All Organisms → Viruses → Duplodnaviria → Heunggongvirae → Uroviricota → Caudoviricetes → environmental samples → uncultured Caudovirales phage | (Source: UniRef50) |
Ecosystem & Geography
| Source Dataset Ecosystem |
|---|
| Engineered → Wastewater → Industrial Wastewater → Petrochemical → Unclassified → Hydrocarbon Resource Environments → Hydrocarbon Resource Environments Microbial Communities From Canada And Usa |
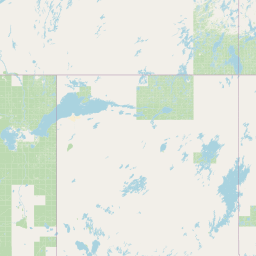
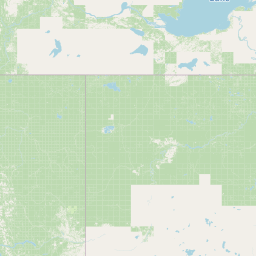
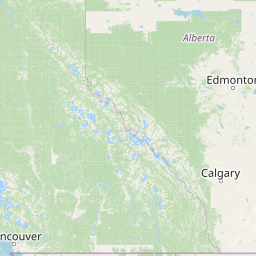
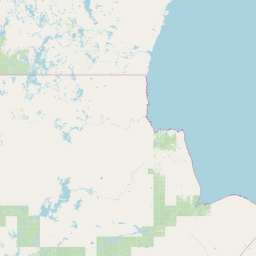
| Source Dataset Sampling Location | ||||||||
|---|---|---|---|---|---|---|---|---|
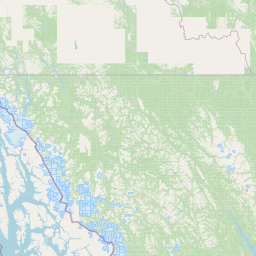
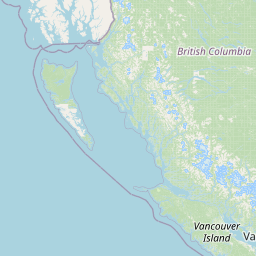
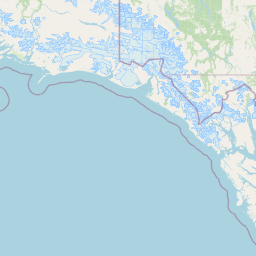
| Location Name | Syncrude, Ft. McMurray, Alberta | |||||||
| Coordinates | Lat. (o) | 57.02 | Long. (o) | -111.55 | Alt. (m) | Depth (m) | ||
| Location on Map | ||||||||
|
| ||||||||
| Zoom: | Powered by OpenStreetMap © | |||||||